SaaS
"DR-Screening" is a solution that supports drug repurposing by quickly discovering new medical uses in selected libraries of 17,000 existing drugs. The libraries include a variety of drugs that are FDA-approved or in preclinical/clinical trials. A proven platform with up to 23.3% hit rates across in various therapeutic areas and multiple protein families, including kinase, GPCR, and nuclear receptors. Furthermore, it is validated to cover various target and disease areas.
Are you considering an ever greening strategy to extend the patent life of your drugs?
STB can help extend the patent rights by adding new indications based on accumulated experiences.
Need to quickly add new pipelines Or find treatment for new pandemic diseases in the fastest way?
STB can support it with drug repurposing with selected DR libraries and integrated technology.
looking for HIT compounds that meet the safety and physicochemical properties?
STB can help reduce development time and costs by finding new uses of existing or investigational drugs.
ADR Library (Drug repurposing)
I/O: Target protein info. & Ref. drug info. (if available)
BDeep Learning Screening
CFalse Positive Filtering
Output: List of 200 Cpds with false positives filtered
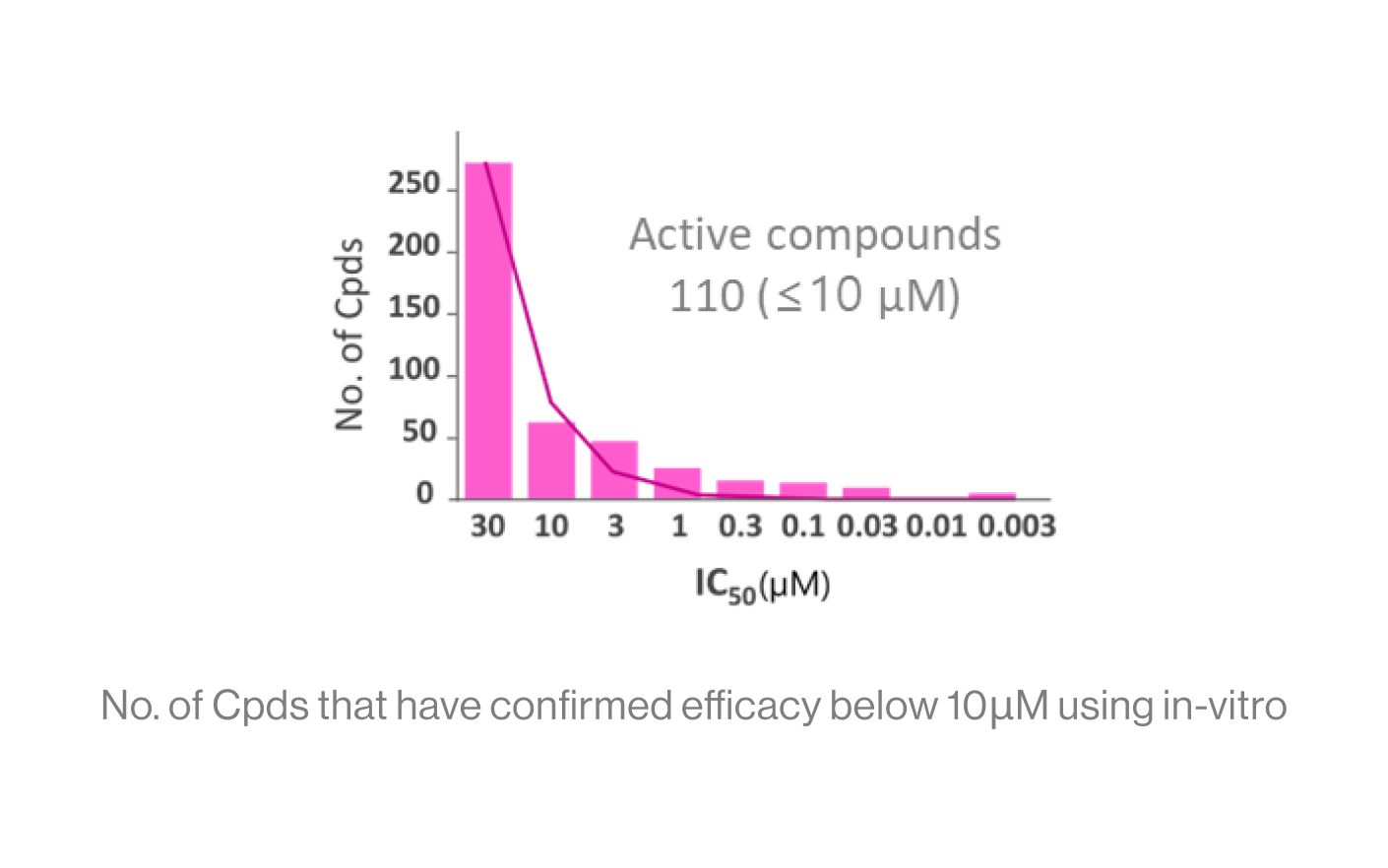
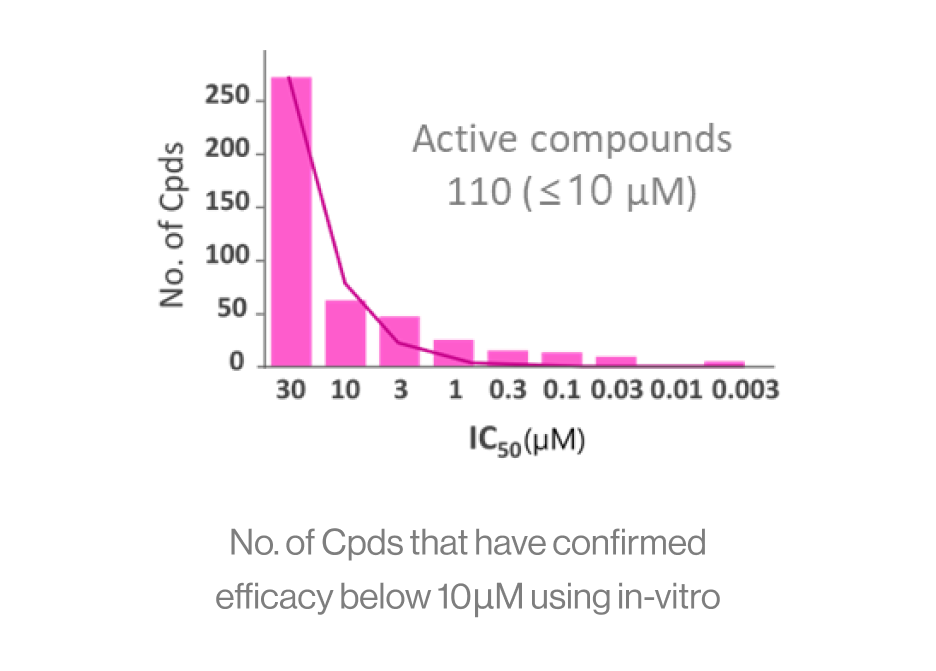
DTechnical Performance
Input: The screening score of 200 Cpds
Auto-Hit Discovery by DeepMatcher®-Hit
Flow diagram of Hit discovery by DeepMatcher® Compounds are prescreened through three steps before being docked in a pocket of the target. Once the optimal pose is selected, the MD simulation is finally performed in a total of five steps.
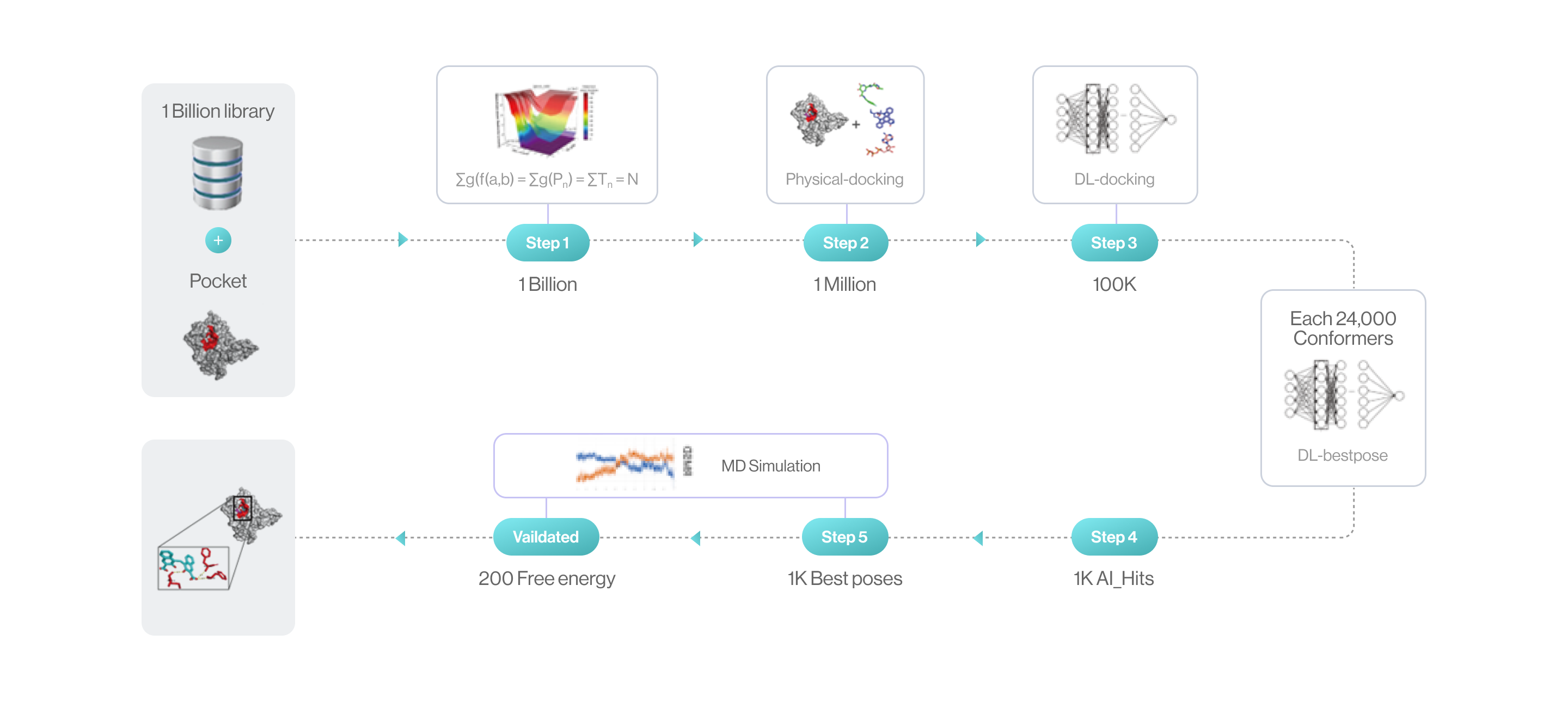
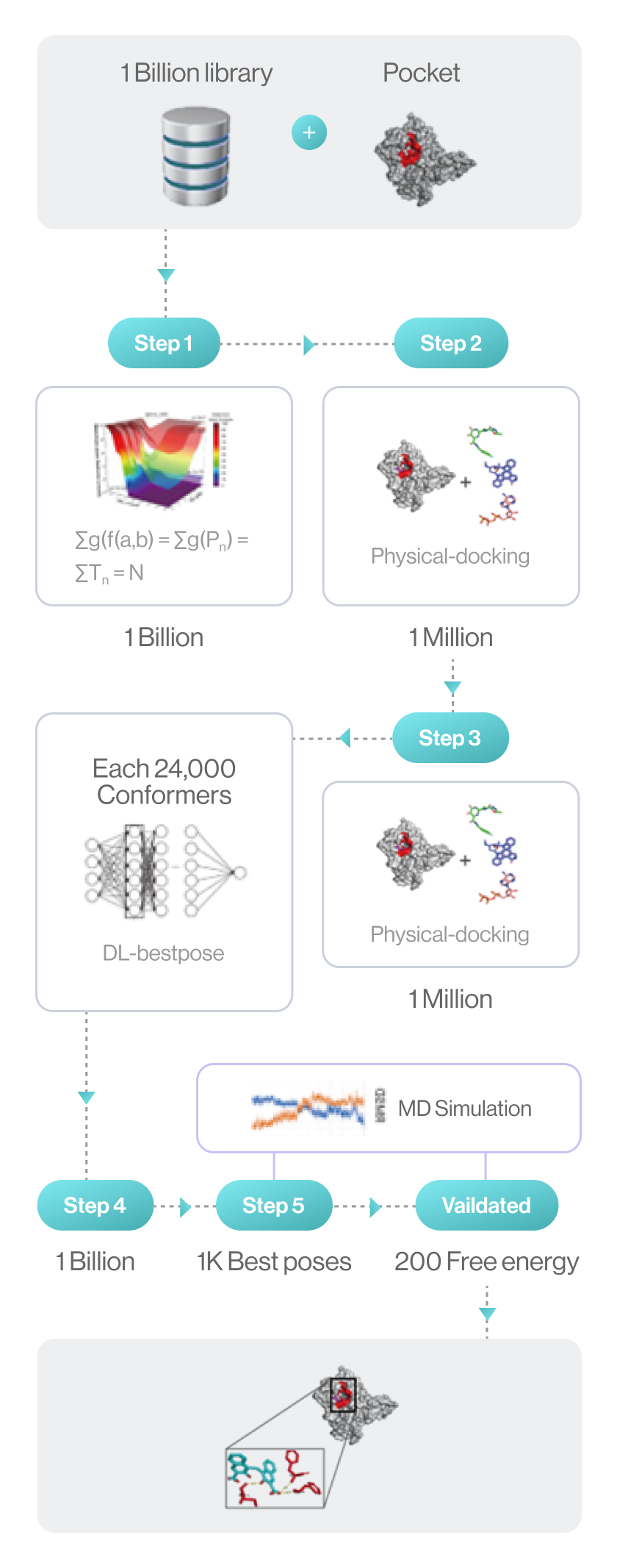
Syntekabio launched STB CLOUD in late 2022, a suite of cloud-based proprietary platforms, to meet the unmet needs of these cost-conscious innovators seeking reliable yet affordable new drug candidate discovery services. STB CLOUD screens 1 billion purchasable compounds based on unique featurization method for 3D information-based data and 1 million potential candidate compounds
One million chemicals selected from a library of 1 billion compounds was docked into target 3D structure using command line interface (CLI) based in-house structure manipulation package.
Figure2. Chemicallibrary
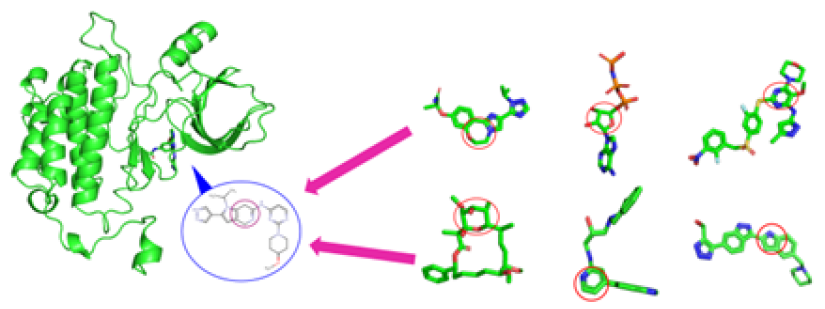
Figure3. Methods for centroidal atom vector-based fast protein-ligand docking. After docking, three atom-vector based rotations along the core ring. It rotates while moving 1000 times in each revolution and then, generates a protein-ligand interaction (PLI) score
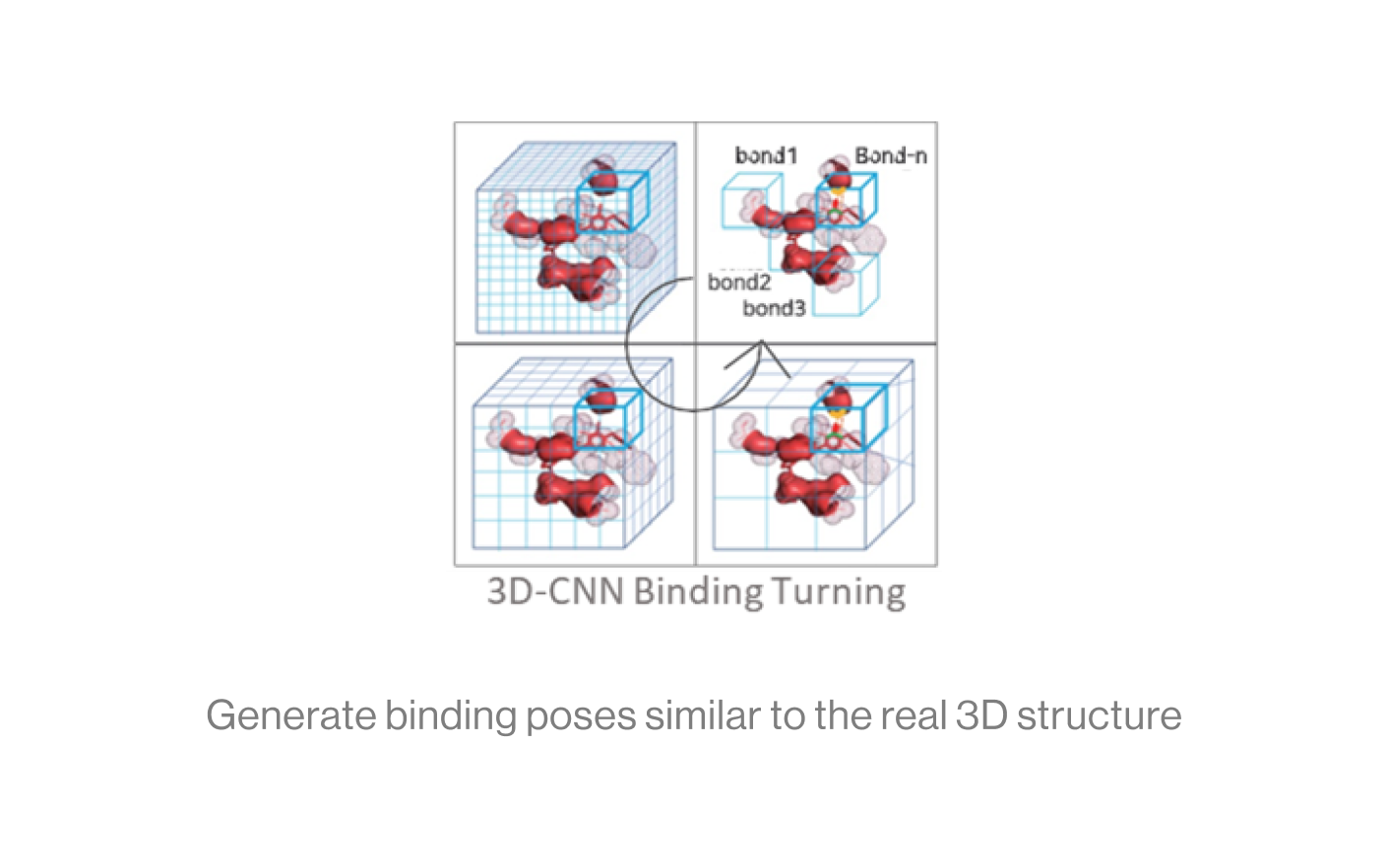
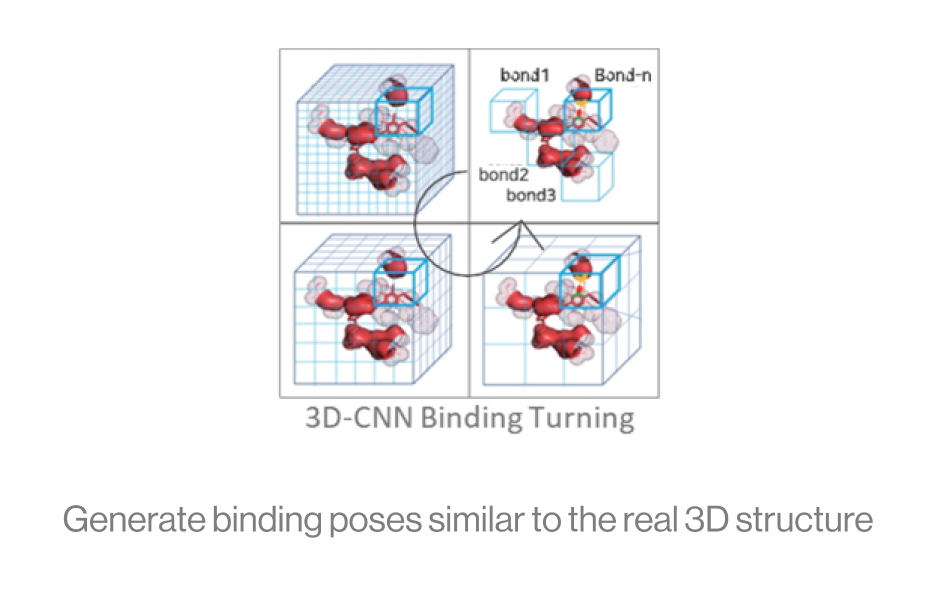
The diagram below shows the interaction between a protein and a ligand in the pocket as a grid box, a 3D-CNN based intermediate stage of repeated max pooling and 3D convolution, generating feature maps, and representing bonds. Here, the starting grid box (48) and kernel size (3*3*3) are used. The density function is based on the geometry of adjacent bonds of two atoms. Therefore, the sum of bonds is defined as binding affinity.
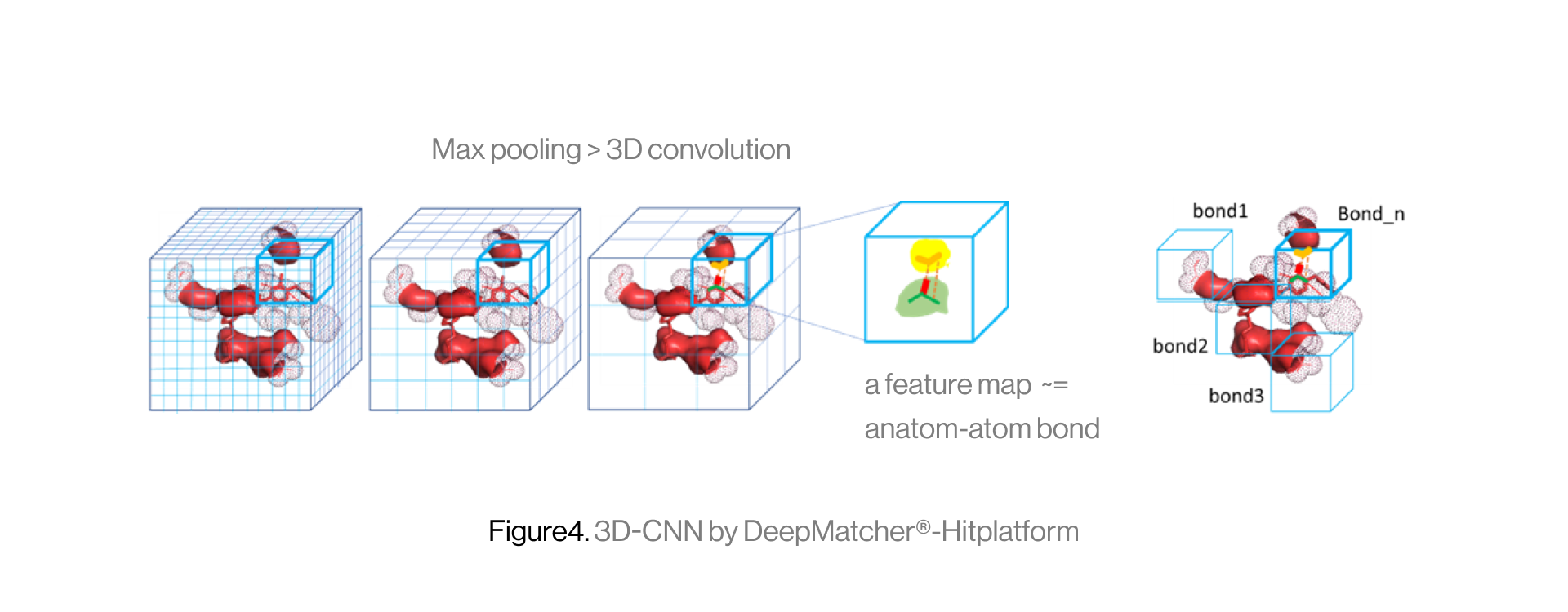
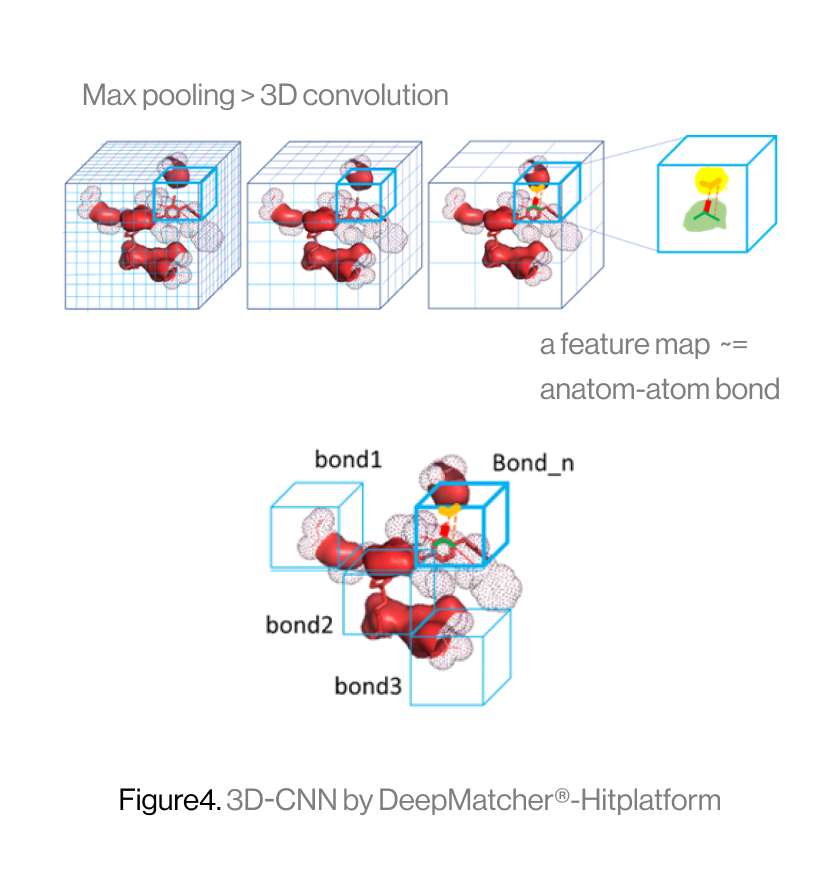
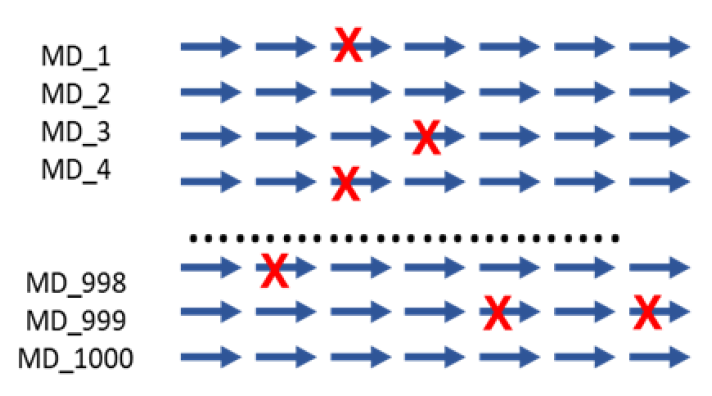
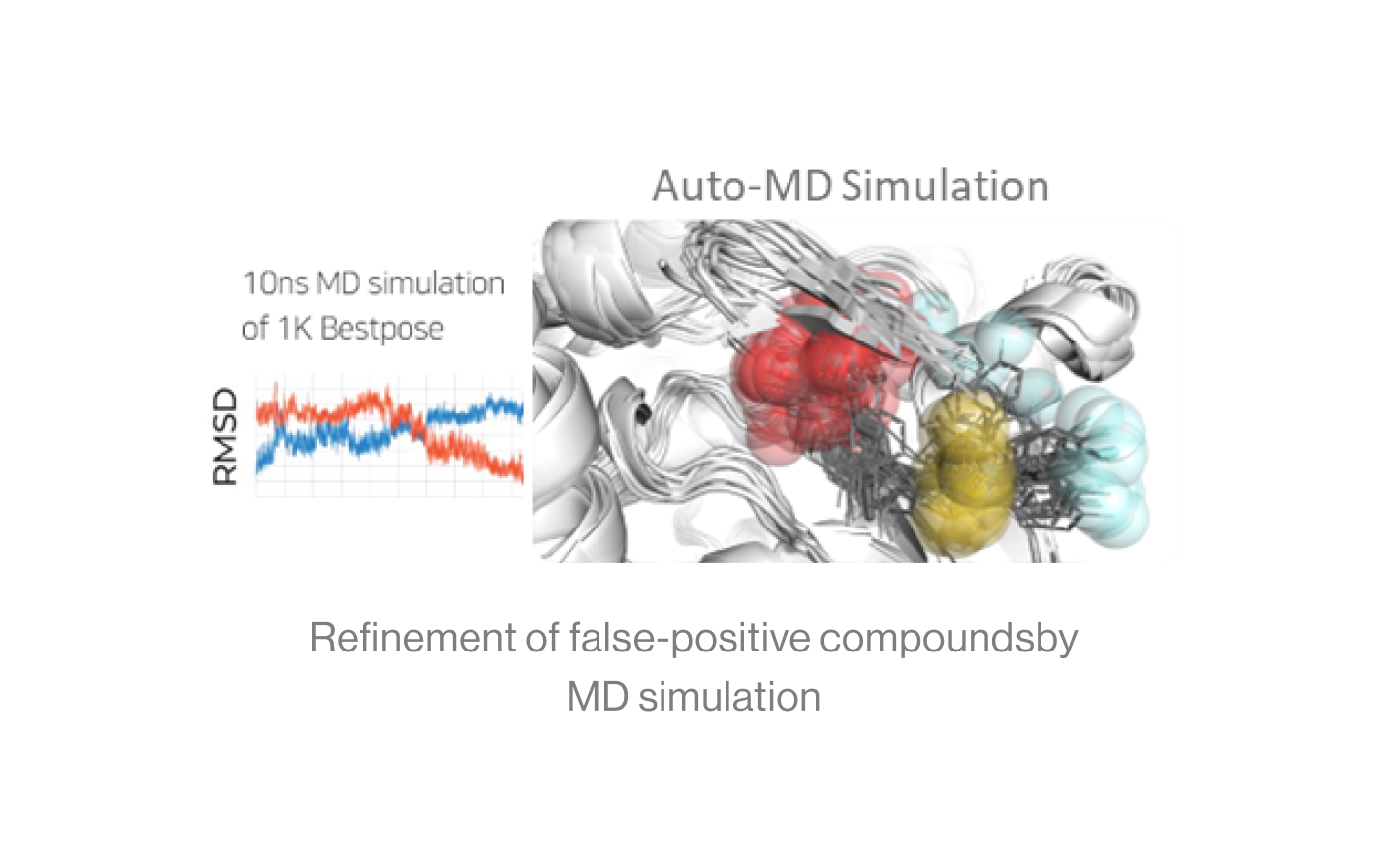
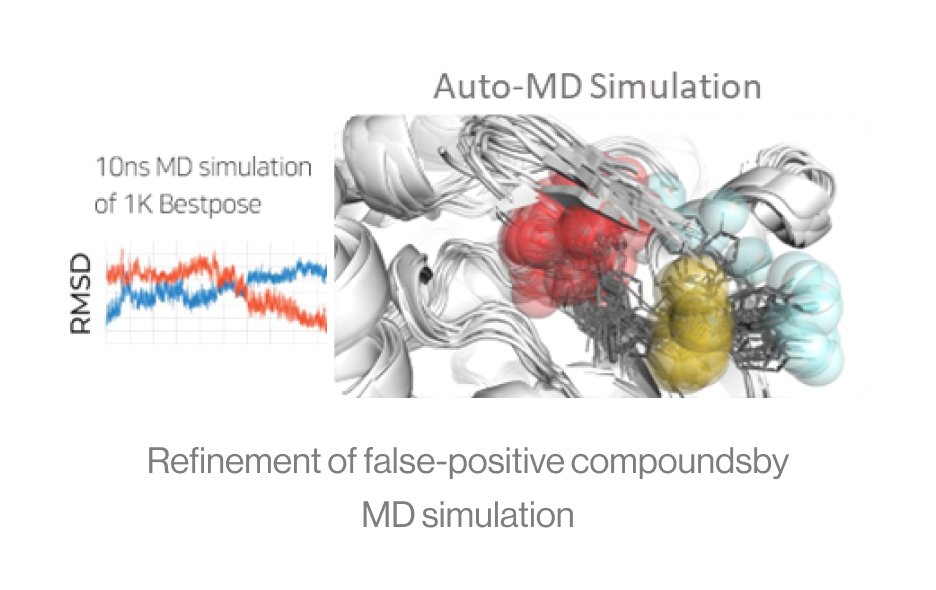
Figure5. Auto-MD simulation
Auto-MD Simulation
Syntekabio's fully automated MD simulation algorithm performs molecular dynamics
simulations (10 ns) in explicit water box for 1,000 protein-ligand complexes.
The diagram shows an example of running continuously from a dead step during 1,000 MD simulations.
If one runs 1000 MD simulations at once, a large number of simulations may crash and die in
certain cases. However, in the automated environment, it proceeds by saving the checkpoint and
resume from the saved point.
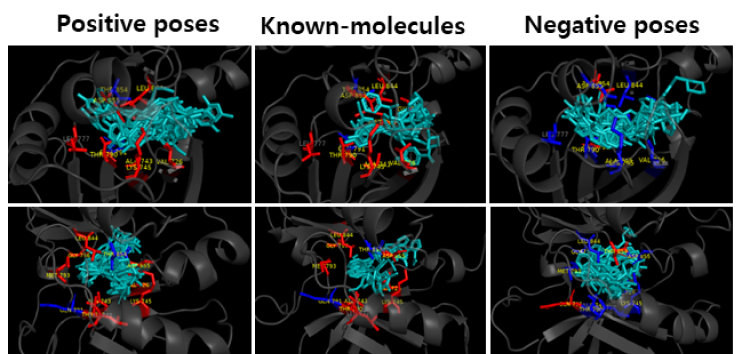
Figure 6. Comparison of AI-hit positive andnegative
predictions with known
Molecules.
Key-Residue Analysis
The known molecules and the predicted molecules with EGFR protein were compared to see if the functional (Hinge, DFG) and conformational keys (G-loop, M766 in c-Helix) were well matched. In the case of positive prediction, the functional and conformational keys were well matched, whereas in case of negative, there were few matched residues. (Red: strong binding, Blue: weak binding)
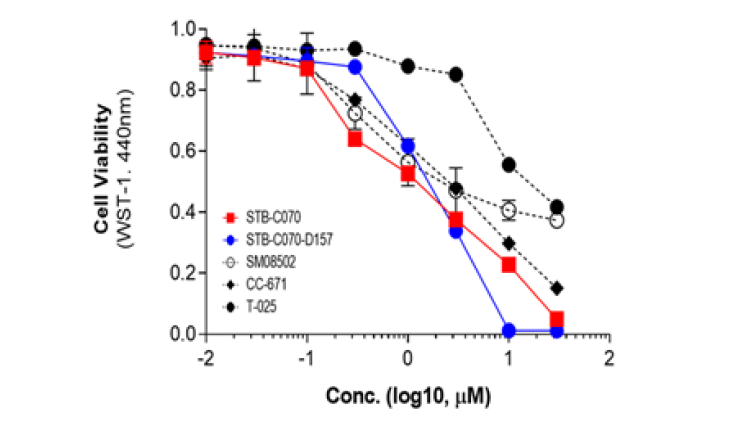
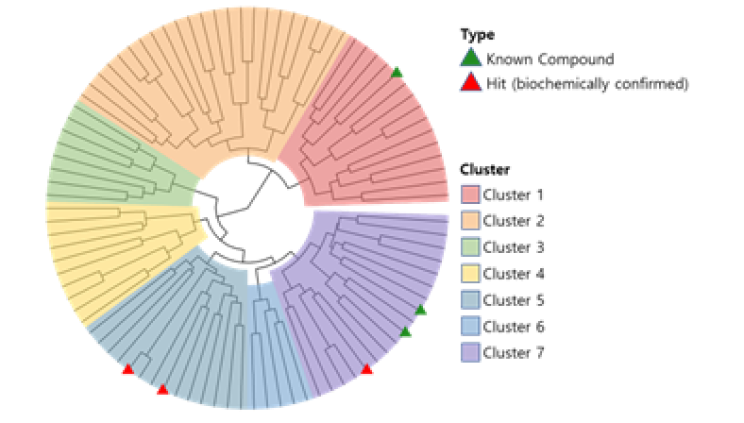
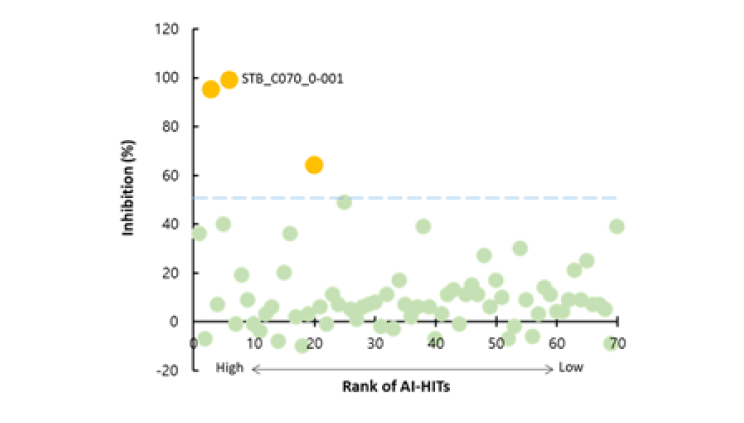
CLK2, also known as CDC-like kinase 2, is a protein kinase that plays a role in the regulation of pre-mRNA splicing. Presented 89 candidate compounds using Deepmatcher®-Hit as a result of structure analysis using tanimoto coefficient with KNN algorithm. a) It was confirmed that 89 compounds were divided into 7 groups. b) Purchasable compounds were biochemically confirmed their inhibition activities. Three compounds of 74 purchased candidates (yellow circles) inhibited enzyme activity of CLK2 more than 50%. c) The growth inhibitory effect of each cancer cell by treatment with STB-C070, STB-C070-D157 and reference compounds were analyzed using WST-1 assay. GI50 was calculated from the 8-point dose-response curves by triplicate test.
a) DeepMatcher® Result analysis
b) Biochemical assay result of AI-hits
c) Cell-based assay result of AI-hits