Automate it by incorporating to STB CLOUD with
in the second quarter of
2023.
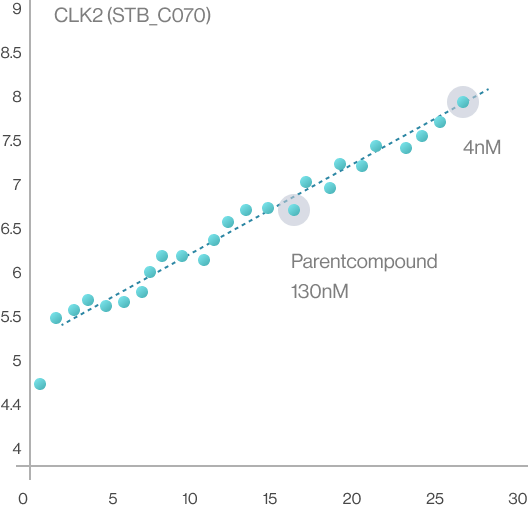
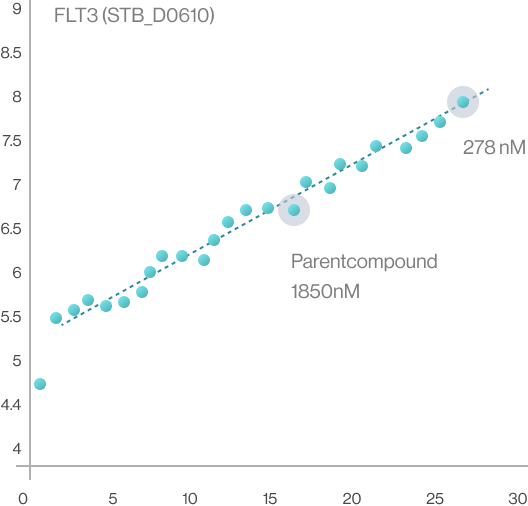
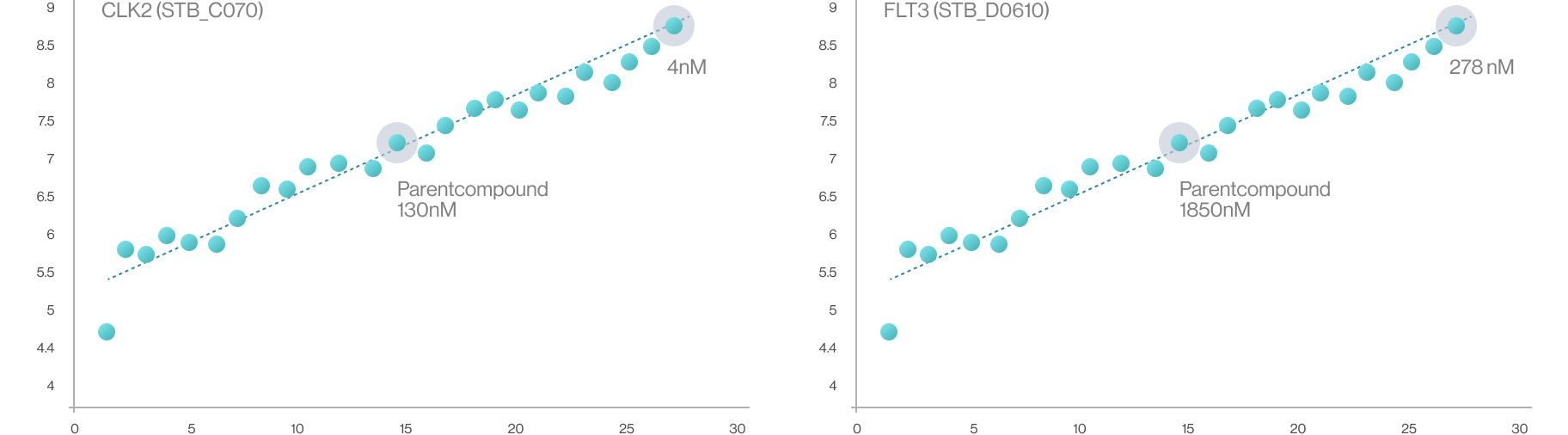
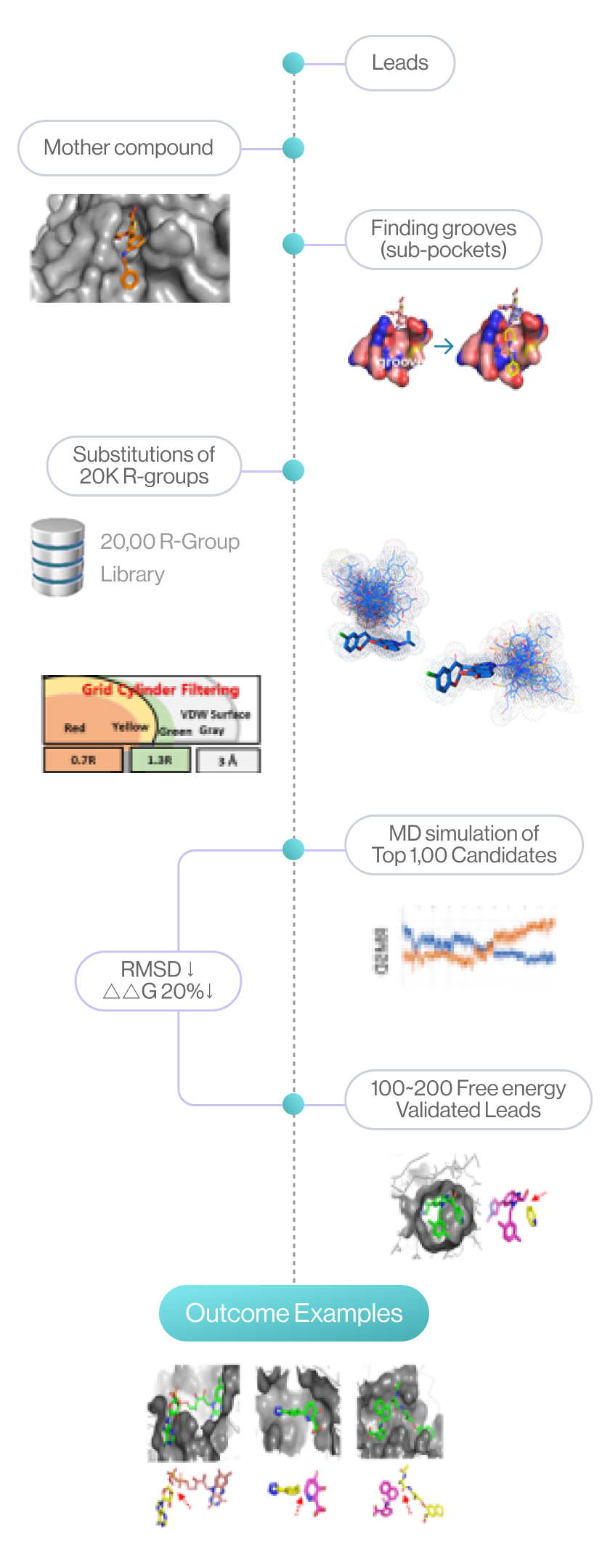
Syntekabio currently has its own library of about 20,000 of R-groups. According to the published works
in this field, the attempts to attach the R-group to the drug scaffold are carried out independently
regardless of the target protein and then brought into the protein’s binding pocket. Contrary to this,
Syntekabio has developed methods to enumerate 20,000 R-groups with high activity directly on the
mother compound within the pocket. Hence, quantitative structure-activity relationship (QSAR) is no
longer necessary as it automatically compares and selects in terms of binding energy. Therefore,
Syntekabio's drug optimization is called Auto-Lead-Optimization. It has been operated
semi-automatically for now in the local environment, with an aim to fully automate it by incorporating
to STB CLOUD within the second quarter of 2023.
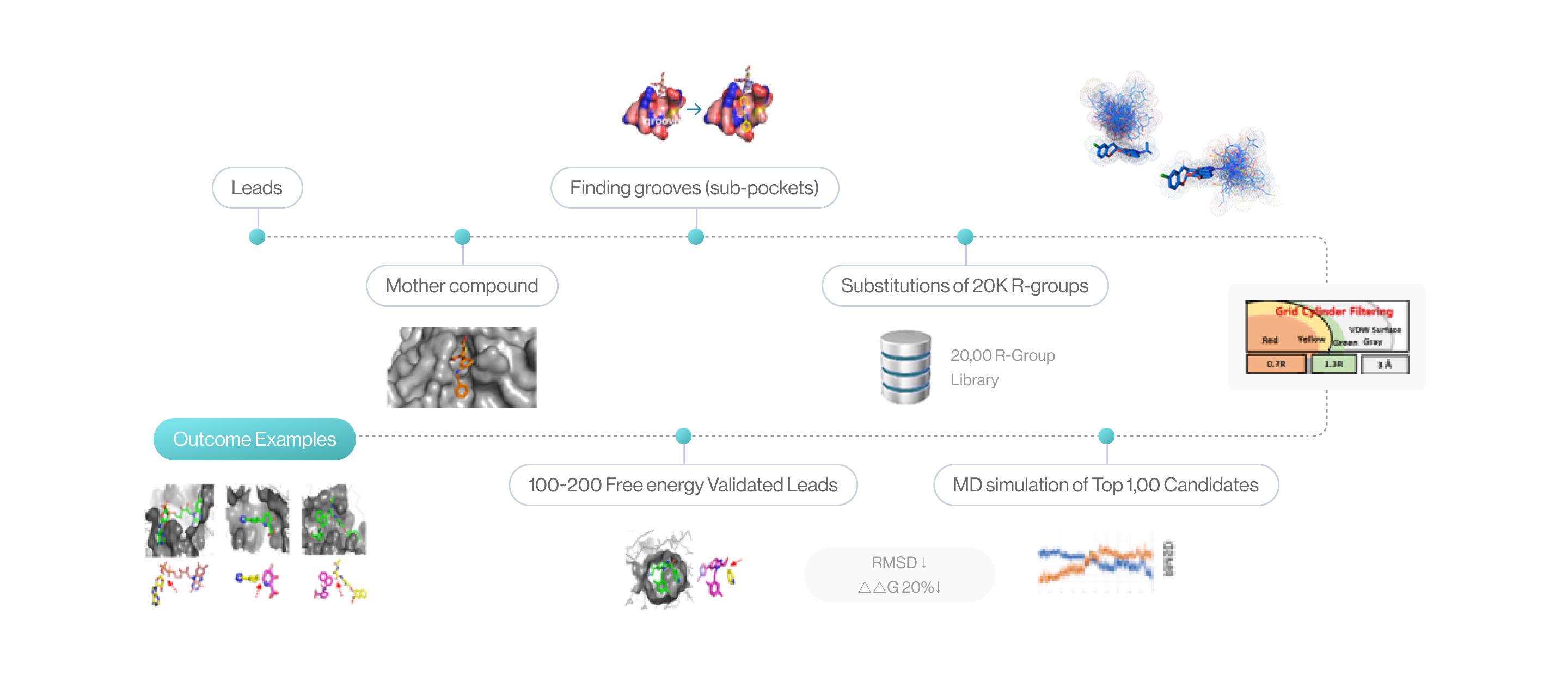
Essential Key-challenges for ALGO(Auto Lead-Gen/Opt)
Flow diagram of Lead generation and optimization by DeepMatcher®-Lead. It is showed R-groups are substituted and perturbed at the position of the mother compound, as the top 1,000 candidates are selected through grid cylinder filtering. The process is finalized through MD simulations.
(source: cloud.syntekabio.com)
1Sub-pocket & Weak Bond Screening
2Substitution of R-group to Scaffold
3Free Energy Perturbation in the Sub-pocket
4MD Simulation for Fine-tune
WeakBond Screening by Scaffold Extraction
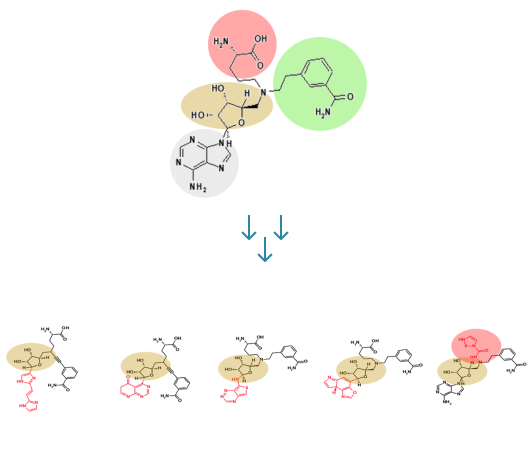
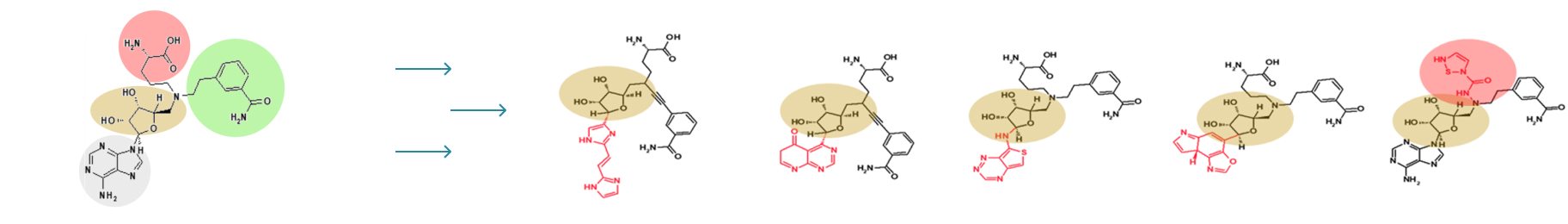
In this step, the weak binding position is found in the optimal binding pose of the hit compound in the pocket. R-group fragments are used at this position to generate derivatives that bind strongly.
While keeping a scaffold from the hit compound in position, we first choose a variety of replaceable regions (red box in the figure) in the hit compound and select one of them which has the lowest interaction efficiency. By removing the regions from the hit compound, we can generate a scaffold. Molecular dynamics (MD) is applied in the same way as shown in hit-discovery. In this step, we select derivatives in the previous step and perform MD simulations.
A
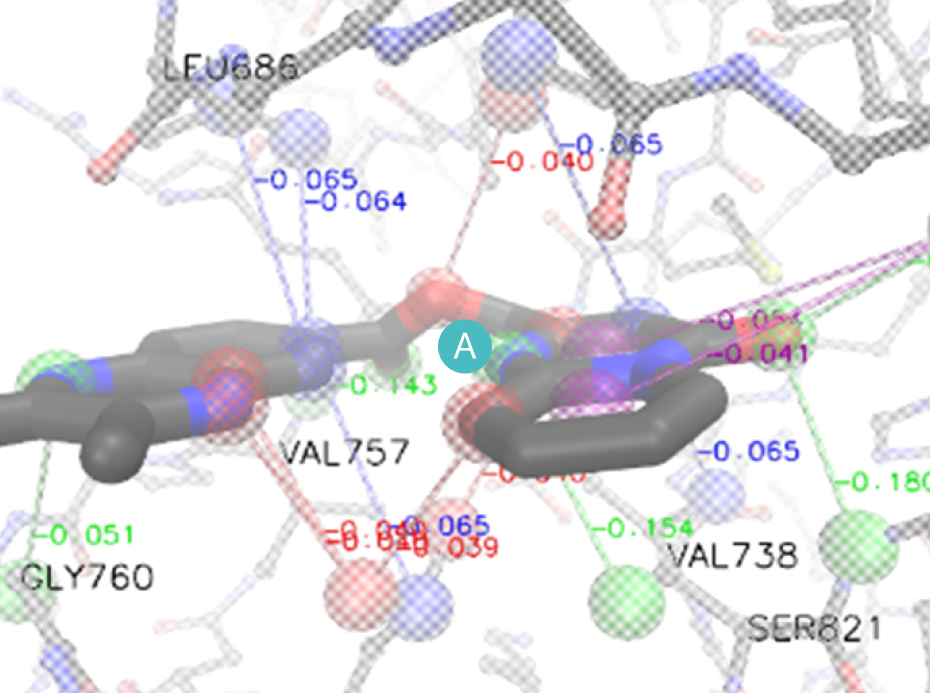
B
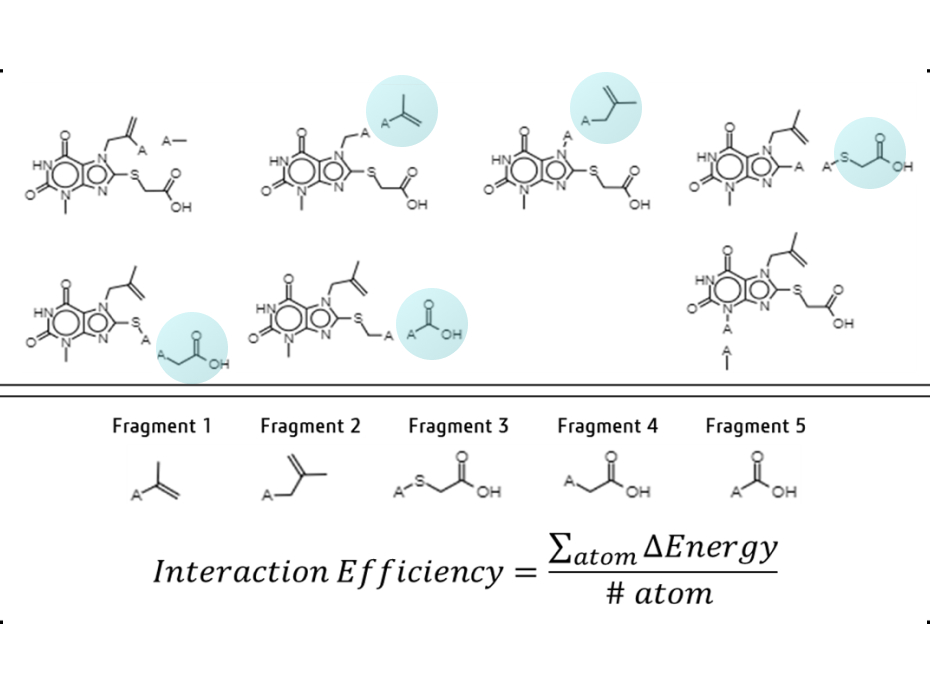
Scaffold extraction from a mother compound.
(A) Binding pose of mother compound, (B) Fragmentation: mother compound interaction
efficiency per atom of each fragment is calculated and substituted for the group with the lowest
B.E. value
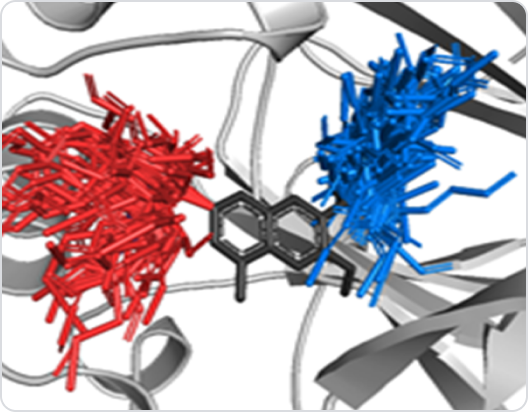
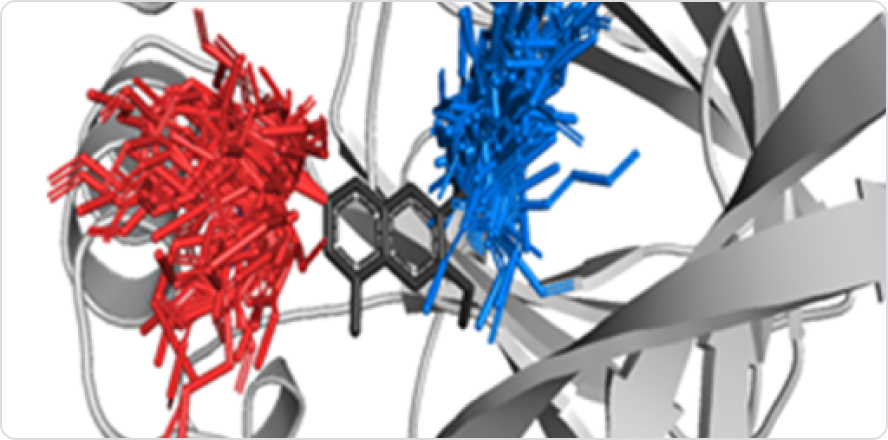
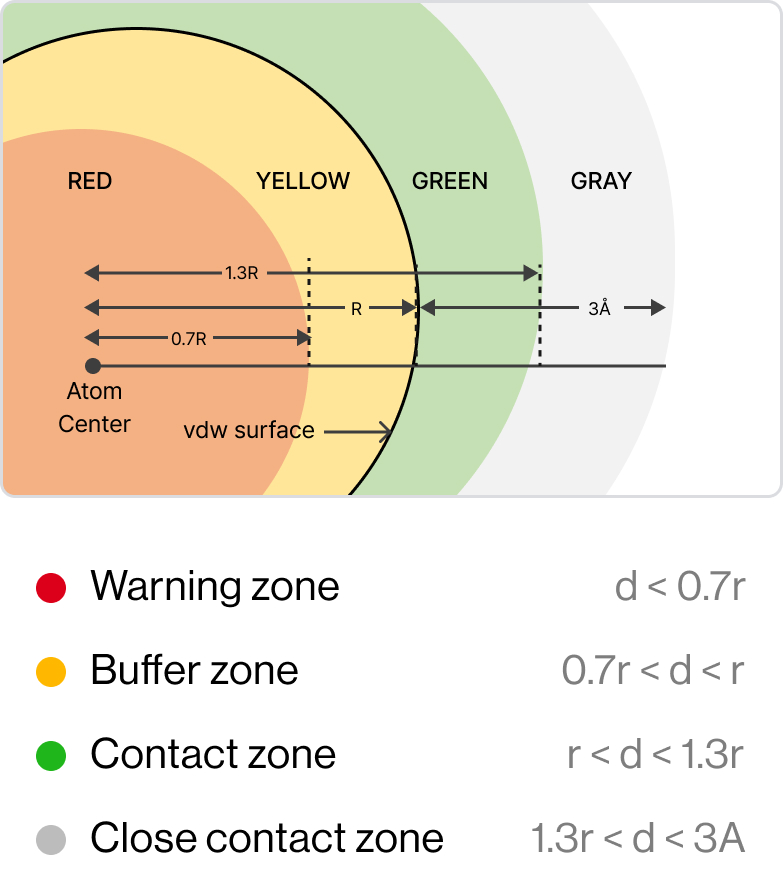
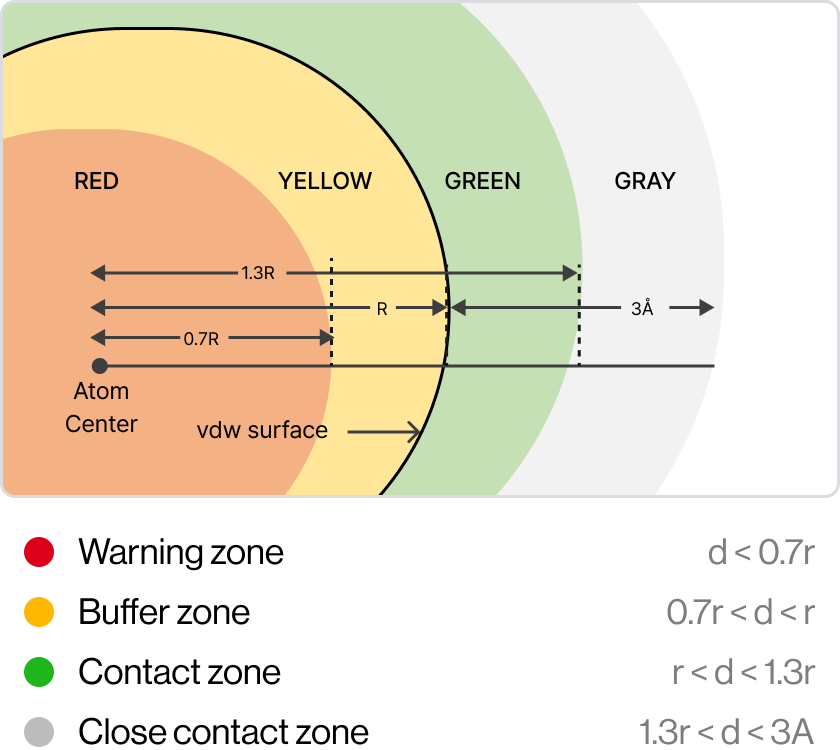
Grid Cylinder Filtering (GCF)
GCFchecks whether new derivatives have reasonable positions in the
pocket without any collisions with theprotein.
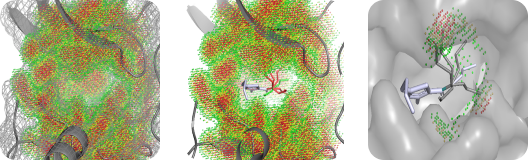
The Diagram shows GCF generates derivatives and filters by properties and shape;
1) Create a filter based on the Van Der Waals radius(r) of each atom, 2) Clash – red / Clash
buffer – yellow / Ideal contact – green / Close contact – Gray, 3) (Rightmost) Blue-white: best
conformation - exactly matched with actual pose
RMSDE valuation of 52 Predicted Poses
Finding the optimal conformation after attaching the R group. Results of
R group attached to the scaffold in known test-set (RMSD with actual binding pose), showing that most
of them are well screened except a few red ones.
RMSD

SMART attach-shape filter
MC_name
avg_rmsd
min_rmsd
count
-
Der-1-1
0.684
0.191
960
-
Der-1-3
1.247
0.242
1020
-
Der-1-4
1.327
0.350
405
-
Der-1-5
1.441
0.268
1042
-
Der-1-6
1.292
0.179
1125
-
Der-1-7
0.702
0.216
787
-
Der-2-1
1.654
0.443
181
-
Der-2-2
1.080
0.721
33
-
Der-2-3
1.604
0.694
25
-
Der-3-1
0.754
0.277
1056
-
Der-3-2
1.235
0.299
1009
-
Der-3-3
1.681
1.475
47
-
Der-3-4
1.001
0.182
284
-
Der-3-5
0.130
0.074
1212
-
Der-4-1
1.208
0.183
900
-
Der-4-10
1.481
0.094
345
-
Der-4-11
0.789
0.234
637
-
Der-4-12
1.028
0.270
490
-
Der-4-3
1.428
0.233
386
-
Der-4-4
1.062
0.285
432
-
Der-4-6
1.206
0.286
370
-
Der-4-7
2.116
0.143
327
-
Der-4-8
0.603
0.183
867
-
Der-4-9
0.128
0.072
1212
-
Der-5-1
1.322
0.759
40
-
Der-5-2
1.755
1.733
3
-
Der-5-3
1.347
0.808
68
-
Der-5-4
1.1.208
0.558
58
-
Der-5-5
1.514
0.685
48
-
Der-5-6
1.276
0.773
88
-
Der-6-1
2.448
0.514
31
-
Der-6-2
3.577
0.803
29
-
Der-7-1
2.305
0.619
315
-
Der-7-2_SN1
2.676
0.840
198
-
Der-7-2_SN2
0.805
0.214
1830
-
Der-7-3_SN1
2.414
0.908
199
-
Der-7-3_SN2
0.828
0.398
960
-
Der-8-1
2.784
0.505
354
-
Der-8-2
3.043
0.911
395
-
Der-8-3
4.347
2.767
268
-
Der-8-4
3.106
1.443
256
-
Der-9-1
9.664
4.341
99
-
Der-9-2_SN1
2.030
0.763
286
-
Der-9-2_SN2
1.953
0.659
96
-
Der-9-3_SN1
0.984
0.053
810
-
Der-9-3_SN2
1.421
1.061
32
-
Der-9-4_SN1
0.921
0.052
720
-
Der-9-4_SN2
1.054
0.321
146
-
Der-9-5_SN1
0.981
0.122
750
-
Der-9-5_SN2
1.468
1.033
31
-
Der-9-6_SN1
0.953
0.063
732
-
Der-9-6_SN2
0.651
0.323
199
SMART attach-shape filter
MC_name
avg_rmsd
min_rmsd
count
-
Der-1-1
0.684
0.191
960
-
Der-1-3
1.247
0.242
1020
-
Der-1-4
1.327
0.350
405
-
Der-1-5
1.441
0.268
1042
-
Der-1-6
1.292
0.179
1125
-
Der-1-7
0.702
0.216
787
-
Der-2-1
1.654
0.443
181
-
Der-2-2
1.080
0.721
33
-
Der-2-3
1.604
0.694
25
-
Der-3-1
0.754
0.277
1056
-
Der-3-2
1.235
0.299
1009
-
Der-3-3
1.681
1.475
47
-
Der-3-4
1.001
0.182
284
-
Der-3-5
0.130
0.074
1212
-
Der-4-1
1.208
0.183
900
-
Der-4-10
1.481
0.094
345
-
Der-4-11
0.789
0.234
637
-
Der-4-12
1.028
0.270
490
-
Der-4-3
1.428
0.233
386
-
Der-4-4
1.062
0.285
432
-
Der-4-6
1.206
0.286
370
-
Der-4-7
2.116
0.143
327
-
Der-4-8
0.603
0.183
867
-
Der-4-9
0.128
0.072
1212
-
Der-5-1
1.322
0.759
40
-
Der-5-2
1.755
1.733
3
SMART attach-shape filter
MC_name
avg_rmsd
min_rmsd
count
-
Der-5-3
1.347
0.808
68
-
Der-5-4
1.1.208
0.558
58
-
Der-5-5
1.514
0.685
48
-
Der-5-6
1.276
0.773
88
-
Der-6-1
2.448
0.514
31
-
Der-6-2
3.577
0.803
29
-
Der-7-1
2.305
0.619
315
-
Der-7-2_SN1
2.676
0.840
198
-
Der-7-2_SN2
0.805
0.214
1830
-
Der-7-3_SN1
2.414
0.908
199
-
Der-7-3_SN2
0.828
0.398
960
-
Der-8-1
2.784
0.505
354
-
Der-8-2
3.043
0.911
395
-
Der-8-3
4.347
2.767
268
-
Der-8-4
3.106
1.443
256
-
Der-9-1
9.664
4.341
99
-
Der-9-2_SN1
2.030
0.763
286
-
Der-9-2_SN2
1.953
0.659
96
-
Der-9-3_SN1
0.984
0.053
810
-
Der-9-3_SN2
1.421
1.061
32
-
Der-9-4_SN1
0.921
0.052
720
-
Der-9-4_SN2
1.054
0.321
146
-
Der-9-5_SN1
0.981
0.122
750
-
Der-9-5_SN2
1.468
1.033
31
-
Der-9-6_SN1
0.953
0.063
732
-
Der-9-6_SN2
0.651
0.323
199